Introduction
The tumour of the testis is one of the most common
type of tumours in the field of urology (1). Testicular tumours are divided into
seminoma and non-seminoma types, the vast majority of which are
seminoma; non-seminoma types are extremely rare. Testicular tumours
are almost all malignant, with germ cell tumours accounting for
90–95%, and non-germ cell tumours accounting for 5–10% (2). In germ cell tumours, seminoma is the
most common, accounting for 40% to 50% of primary testicular
tumours, followed by embryonic cancer (20–30%) and teratoma (~10%)
(3–5). Left and right testicular tumours of
other cell types are rare. The treatment of testicular tumours is
divided into single treatment, and the comprehensive treatment of
surgical treatment radiation therapy and chemotherapy (6). Unfortunately, once a testicular
tumour is identified, radical orchiectomy should be performed
first, and then a further treatment plan should be implemented
based on the pathological findings (7–10).
Hence, it is urgent and necessary to explore novel therapeutic
targets for the treatment of seminoma.
In the present study, we selected three gene
expression datasets (GSE15220, GSE1818 and GSE59520), which were
downloaded from the Gene Expression Omnibus (GEO) database, to
obtain differentially expressed genes (DEGs) and differentially
expressed microRNAs (DEMs) between testicular seminoma tissues and
normal tissue samples. Then, functional enrichment and network
analyses were applied to identify the DEGs. Subsequently, we
established a protein-protein interaction (PPI) network to identify
hub genes related to seminoma. The expression values of these hub
genes were determined using the online database UALCAN. Survival
analysis of these hub genes was performed using the online database
Gene Expression Profiling Interactive Analysis (GEPIA). The
potential target genes of the miRNAs were predicted by miRwalk 3.0
and screened by The Cancer Genome Atlas (TCGA) dataset.
Materials and methods
Identification of DEGs and DEMs
Three gene expression profiles, GSE15220, GSE1818
and GSE59520, were acquired from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The array
data of GSE15220 comprising 6 paired seminoma tissues, and adjacent
tissues were submitted by Cheung et al (11); GSE1818 consisted of 6 paired
seminoma tissues and adjacent tissues. GSE59520 consisted of 14
seminoma tissues and three normal tissues (12). DEGs were obtained from the GEO
database by GEO2R analysis (http://www.ncbi.nlm.nih.gov/geo/geo2r/). Adjusted
P<0.05 and log fold-change (|logFC|) >2.0 were set as the DEG
cutoff criterion. Adjusted P<0.05 and |logFC| >1.0 were set
as the DEM cutoff criterion. The common dysregulated genes between
GSE1818 and GSE59520 are presented as a Venn diagram and identified
using R (version 3.6.1; http://www.r-project.org/). The Search Tool for the
Retrieval of Interacting Genes (STRING) database (https://string-db.org/) for annotation, visualization
and integrated discovery was employed to facilitate the transition
from data collection to biological analysis. Gene Ontology (GO) and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analyses were performed using the Enrichr online tool (http://amp.pharm.mssm.edu/Enrichr/). P<0.05
was set as the cut off criterion. Potential target genes of the
miRNAs were predicted by miRwalk 3.0 (http://mirwalk.umm.uni-heidelberg.de/) and screened
using the TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
database.
GO and KEGG pathway analyses of
DEGs
GO analysis is a commonly used method for
large-scale functional enrichment research; gene functions can be
classified into biological process (BP), molecular function (MF)
and cellular component (CC). KEGG is a widely used database that
stores data on genomes, biological pathways, diseases, chemical
substances and drugs. GO annotation and KEGG pathway enrichment
analyses of the DEGs identified in this study were performed using
Enrichr tools. P<0.05 was considered to indicate a statistically
significant difference.
Integration of the PPI network and hub
gene selection
The STRING database is designed to analyse PPI
information. To evaluate the potential PPI relationships, the DEGs
we identified were mapped to the STRING database. The PPI pairs
with a combined score of 0.4 were extracted. Subsequently, the PPI
network was visualized by Cytoscape software (version 3.7.1;
www.cytoscape.org/). Nodes with a higher degree
of connectivity tend to be more essential in maintaining the
stability of the entire network. CytoHubba (version 0.1) (13), a plugin in Cytoscape, was used to
calculate the degree of each protein node. In our study, the top
five genes were identified as hub genes. miRwalk (version 3.0;
http://mirwalk.umm.uni-heidelberg.de/) was used to
predict the potential target genes of the miRNAs identified.
Expression profiles of hub genes based
on tumour histology and survival analysis
UALCAN (http://UALCAN.path.uab.edu) is a user-friendly,
interactive web resource for analysing cancer transcriptome data.
According to the median expression of a particular gene, the
patients with testicular germ cell tumors (TGCT) were split into
high and low expression groups. The overall survival (OS) of TGCT
patients was evaluated using GEPIA (14). P<0.05 was considered to indicate
a statistically significant result.
Results
Identification of DEGs and DEMs
The gene expression profiles GSE15220, GSE1818 and
GSE59520 were selected in this study. Based on the criteria of
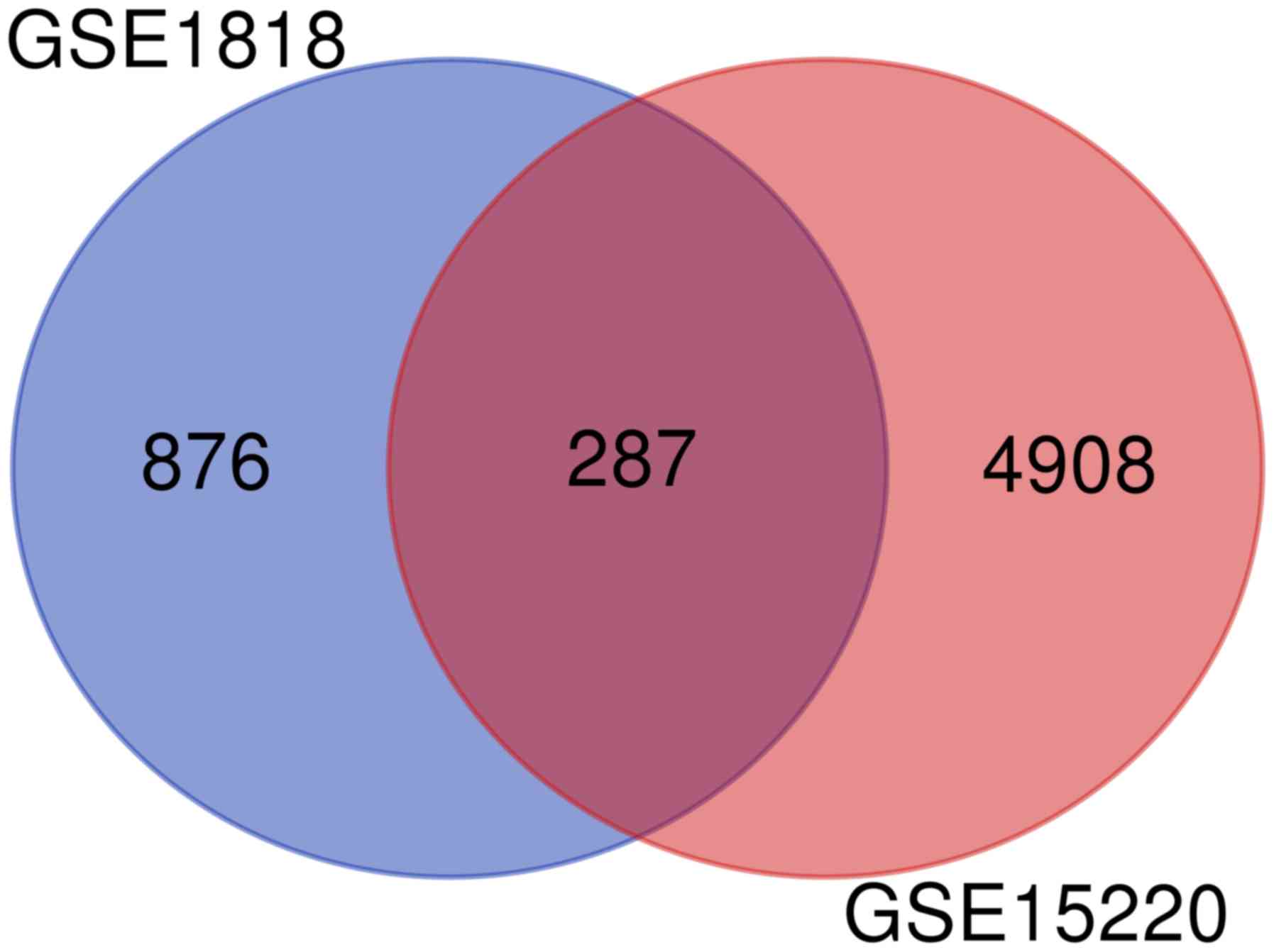
P<0.05 and |logFC|>2.0, a total of 5,195 DEGs were identified
from GSE15220, and 1,163 DEGS were identified from GSE1818. Among
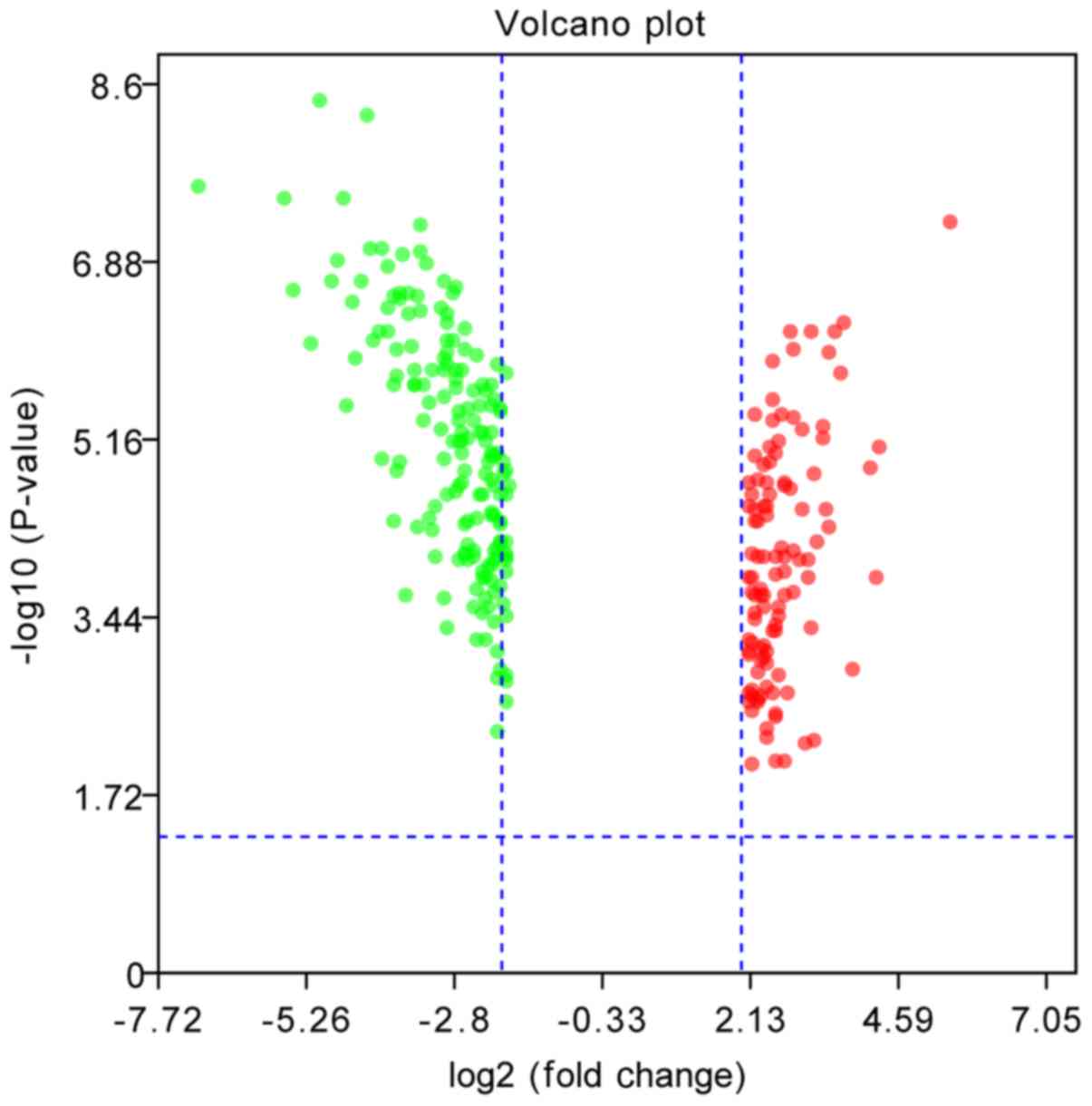
them, 287 genes were common to the two datasets (Fig. 1). Of these, 110 were upregulated,
and 176 were downregulated (Fig.
2). A total of 8 DEMs that are downregulated in seminoma were
identified from GSE59520. All DEGs and DEMs were identified by
comparing seminoma samples with normal samples.
Functional and pathway enrichment
analyses
GO function and KEGG pathway enrichment analyses for
the DEGs were performed using Enrichr (Table I). The enriched GO terms were
divided into CC, BP, and MF ontology terms. The results of GO
analysis indicated that upregulated genes were mainly enriched in
BPs, including ‘regulation of the cellular macromolecule
biosynthetic process’, ‘B cell activation’, ‘regulation of nucleic
acid-templated transcription’, and ‘regulation of gene expression’,
while the downregulated genes were mainly involved in
‘calcium-dependent cell-cell adhesion via plasma membrane cell
adhesion molecules (CAMs)’, ‘spermatid development’,
‘spermatogenesis’, and ‘cell-cell adhesion via plasma membrane
adhesion molecules’. MF analysis showed that the upregulated genes
were significantly enriched in ‘RNA binding’, ‘protein
homodimerization activity’, ‘protein heterodimerization activity’,
and ‘transcription regulatory region DNA binding; downregulated
genes were mainly enriched in ‘motor activity, microtubule motor
activity’, ‘actin binding’, and ‘ATPase activity’. For CC ontology,
the upregulated genes were mainly enriched in ‘focal adhesion’,
‘phagocytic vesicle membrane’, ‘integral component of the luminal
side of the endoplasmic reticulum membrane’, and ‘early endosome
membrane’, and the downregulated genes were mainly enriched in
‘condensed nuclear chromosome’, ‘centromeric region’, ‘cytoplasmic
dynein complex’, ‘spindle midzone’, and ‘focal adhesion’.
 | Table I.Significantly enriched Go terms and
KEGG pathways of differentially expressed genes. |
Table I.
Significantly enriched Go terms and
KEGG pathways of differentially expressed genes.
| A, Upregulated |
|---|
|
|---|
| Category | Term | Description | Count | P-value |
|---|
| BP term | GO:2000112 | Regulation of
cellular macromolecule biosynthetic process | 13 | 0.000044 |
| BP term | GO: 0042113 | B cell activation
(GO:0042113) | 4 | 0.000741 |
| BP term | GO:1903506 | Regulation of
nucleic acid-templated transcription | 12 | 0.000130 |
| BP term | GO:0010468 | Regulation of gene
expression | 15 | 0.000557 |
| CC term | GO:0005925 | Focal adhesion | 8 | 0.000804 |
| CC term | GO:0030670 | Phagocytic vesicle
membrane | 4 | 0.000061 |
| CC term | GO:0071556 | Integral component
of luminal side of endoplasmic reticulum membrane | 3 | 0.000590 |
| CC term | GO:0031901 | Early endosome
membrane | 4 | 0.000741 |
| MF term | GO:0042803 | Protein
homodimerization activity | 16 |
7.14×107 |
| MF term | GO:0003723 | RNA binding | 18 | 0.000550 |
| MF term | GO:0046982 | Protein
heterodimerization activity | 7 | 0.000675 |
| MF term | GO:0044212 | Transcription
regulatory region DNA binding | 10 | 0.003244 |
| KEGG pathway | hsa05230 | Viral
carcinogenesis | 4 | 0.026682 |
| KEGG pathway | hsa04670 | Leukocyte
transendothelial migration | 3 | 0.027385 |
| KEGG pathway | hsa04514 | Cell adhesion
molecules | 6 | 0.000135 |
|
| B,
Downregulated |
|
|
Category | Term |
Description | Count | P-value |
|
| BP term | GO:0016339 | Calcium-dependent
cell-cell adhesion via plasma membrane cell adhesion molecules | 4 | 0.000500 |
| BP term | GO:0007286 | Spermatid
development | 3 | 0.004178 |
| BP term | GO:0007283 |
Spermatogenesis | 4 | 0.047347 |
| BP term | GO:0098742 | Cell-cell adhesion
via plasma-membrane adhesion molecules | 5 | 0.008733 |
| CC term | GO:0000780 | Condensed nuclear
chromosome, centromeric region | 2 | 0.005635 |
| CC term | GO:0005868 | Cytoplasmic dynein
complex | 2 | 0.015846 |
| CC term | GO:0051233 | Spindle
midzone | 2 | 0.030298 |
| CC term | GO:0005925 | Focal adhesion | 7 | 0.038965 |
| MF term | GO:0003774 | Motor activity | 6 | 0.000112 |
| MF term | GO:0003777 | Microtubule motor
activity | 4 | 0.002046 |
| MF term | GO:0003779 | Actin binding | 6 | 0.025579 |
| MF term | GO:0016887 | ATPase
activity | 5 | 0.034558 |
| KEGG pathway | hsa00531 | Amphetamine
addiction | 4 | 0.002886 |
| KEGG pathway | hsa04720 | Long-term
potentiation | 3 | 0.020452 |
| KEGG pathway | hsa04114 | Oocyte meiosis in
diabetic complication | 4 | 0.023439 |
In addition, KEGG pathway analysis showed that the
DEGs were mainly enriched in ‘viral carcinogenesis’, ‘leukocyte
transendothelial migration’ and ‘CAMs’, while the downregulated
genes were mainly enriched in ‘amphetamine addiction’, ‘long-term
potentiation’, and ‘oocyte meiosis in diabetic complications’.
PPI network construction and the
analysis of hub genes
A total of 123 nodes and 269 edges were mapped in
the PPI network of the identified DEGs (Fig. 3). The 5 nodes with the highest
degrees, including DNA topoisomerase II α (TOP2A), ubiquitin
conjugating enzyme E2 C (UBE2C), protein tyrosine
phosphatase receptor type C (PTPRC), marker of proliferation
Ki-67 (MKI67), and centromere protein A (CENPA), were
screened as hub genes (Fig. 4,
Table II). GO term enrichment
analysis showed that in BPs, the genes in this module were mainly
associated with ‘negative regulation of chromosome organization’,
‘regulation of metaphase/anaphase transition of cell cycle’,
‘regulation of cell cycle process’, and ‘condensed nuclear
chromosome kinetochore’ (Table
III). The genes were significantly enriched in the ‘condensed
nuclear chromosome kinetochore’, ‘nuclear ubiquitin ligase complex’
‘chromosome’, and ‘nucleolus’ (Table
III). MF analysis showed that the genes were mainly enriched in
‘ubiquitin-like protein conjugating enzyme activity’, ‘protein
kinase binding’, and ‘ubiquitin binding’ (Table III). KEGG analysis revealed that
the genes were mainly enriched in ‘cell adhesion molecules’,
‘primary immunodeficiency’, and ‘Fc gamma R-mediated phagocytosis’
(Table III).
 | Table II.Top five hub genes with the highest
degrees of connectivity. |
Table II.
Top five hub genes with the highest
degrees of connectivity.
| Gene | Gene
description | Degree |
|---|
| TOP2A | Topoisomerase (DNA)
II α 170 kDa | 33 |
| UBE2C |
Ubiquitin-conjugating enzyme E2C | 16 |
| PTPRC | Protein tyrosine
phosphatase, receptor type, C | 15 |
| MKI67 | Antigen identified
by monoclonal antibody Ki-67 | 13 |
| CENPA | Centromere protein
A | 13 |
 | Table III.Significantly enriched GO terms and
KEGG pathways of the top five hub genes. |
Table III.
Significantly enriched GO terms and
KEGG pathways of the top five hub genes.
| Category | Term | Description | Count | P-value |
|---|
| BP term | GO:2001251 | Negative regulation
of chromosome organization | 1 | 0.001749 |
| BP term | GO:1902099 | Regulation of
metaphase/anaphase transition of cell cycle | 1 | 0.002248 |
| BP term | GO:0010564 | Regulation of cell
cycle process | 1 | 0.022500 |
| CC term | GO:0000778 | Condensed nuclear
chromosome kinetochore | 2 | 0.002997 |
| CC term | GO:0005730 | Nucleolus | 2 | 0.010690 |
| CC term | GO:0005694 | Chromosome | 2 | 0.000235 |
| CC term | GO:0000152 | Nuclear ubiquitin
ligase complex | 1 | 0.010460 |
| MF term | GO:0061650 | Ubiquitin-like
protein conjugating enzyme activity | 1 | 0.005488 |
| MF term | GO:0019198 | Transmembrane
receptor protein phosphatase activity | 1 | 0.003994 |
| MF term | GO:0043130 | Ubiquitin
binding | 1 | 0.015650 |
| MF term | GO:0019901 | Protein kinase
binding | 2 | 0.005841 |
| KEGG pathway | hsa04514 | Cell adhesion
molecules | 1 | 0.035000 |
| KEGG pathway | hsa05340 | Primary
immunodeficiency | 1 | 0.009217 |
| KEGG pathway | hsa04666 | Fc gamma R-mediated
phagocytosis | 1 | 0.023040 |
Expression profiles of the hub genes
and survival analysis
To investigate the expression and prognostic values
of the five potential hub genes, the UALCAN bioinformatics analysis
platform was used. All of the hub genes were significant (Fig. 5). We found that the high expression
of these hub genes was associated with an unfavourable OS of
patients with testicular seminoma by GEPIA (Fig. 6). However, because of the better
prognosis of testicular cancer and few mortalities, the
overexpression of only PTPRC was identified as an
unfavourable prognostic overall survival in patients with
seminoma.
 | Figure 5.Expression values of the top five
DEGs in seminoma and non-seminoma tissues. The horizontal lines in
the figure represent maximum, upper quartile, median, lower
quartile, minimum from top to bottom. CENPA, centromere protein A;
MKI67, marker of proliferation Ki-67; PTPRC, protein tyrosine
phosphatase receptor type C; TGCT, testicular germ cell tumor;
TOP2A, DNA topoisomerase II α; UBE2C, ubiquitin conjugating enzyme
E2 C. |
 | Figure 6.Gene Expression Profiling Interactive
Analysis for overall survival associated with the expression of the
five hub genes in patients with testicular cancer. Red line
represents high expression, and blue line represents low
expression. CENPA, centromere protein A; HR, hazard ratio; MKI67,
marker of proliferation Ki-67; n, number of samples; PPI,
protein-protein interaction; PTPRC, protein tyrosine phosphatase
receptor type C; TPM, transcripts per million; TOP2A, DNA
topoisomerase II α; UBE2C, ubiquitin conjugating enzyme E2 C. |
Potential target genes of the
miRNAs
To predict the potential target genes of the miRNAs,
miRwalk and Cytoscape were used. miR-661 was found to be associated
with PTPRC, miR-640 and miR-665 to MKI67, miR-1204 with CENPA,
miR-1203 with UBE2C, miR-650 and 934 with TOP2A, and miR-1182 with
TOP2A, UBE2C and MKI67 (Fig.
7).
Discussion
Although the cure and survival rates of testicular
cancer are high, the main treatment methods are chemotherapy and
resection (15), which presents a
great burden on the patient quality of life. In addition, the
incidence of seminoma is rising. The lifestyles of males in
different regions vary, while the incidence rate also differs
(16). It is estimated that by
2030, there will be 65,827 new cases worldwide, an increase of
10,561 cases from 2012 (17).
Although multiple approaches have reduced mortality, treatment
resistance, disease relapse and treatment-derived side effects in
particular, are important issues at present (18). Therefore, it is important to
develop novel specific targeted therapies for the treatment of
seminoma.
In our study, a total of 287 DEGs were screened,
including 110 upregulated genes and 177 downregulated genes. The
upregulated genes were enriched in ‘viral carcinogenesis’,
‘leukocyte transendothelial migration’ and ‘CAMs’, while the
downregulated genes were mainly enriched in ‘amphetamine
addiction’, ‘long-term potentiation’ and ‘oocyte meiosis in
diabetic complications’. Moreover, by constructing the PPI network,
we identified five high-degree hub genes, including TOP2A,
UBE2C, PTPRC, MKI67 and CENPA. Among them, all hub genes
were upregulated in seminoma. Finally, we used UALCAN to analyse
the expression of these hub genes; all the hub genes were reported
to be significant. Then, we predicted the association between the
expression of the hub genes and the prognosis of TGCT patients.
Based on GEPIA, the overexpression of all hub genes was related to
an unfavourable prognosis in patients with testicular cancer. Among
them, we found that the overexpression of PTPRC was an unfavourable
prognostic factor for patients with seminoma.
TOP2A is the molecular target of several clinically
useful chemotherapeutic drugs and has been used to treat a variety
of tumours, including breast cancer, prostate cancer and
endometrial cancer (19). Certain
studies have suggested that the overexpression of TOP2A may
be associated with the poor prognosis of these malignant diseases.
For seminoma, TOP2A overexpression was associated with
aggressive clinical behaviours (20). Coleman et al (21) and Sano and Shuhin (22) suggested that TOP2A may be a
marker of seminoma cell proliferation, and TOP2A was easily
detected in seminoma. Additionally, studies have also revealed that
TOP2A is related to primary tumours (23–25).
In prostate cancer, Labbé et al (26) found that TOP2A and EZH2 mRNA
and protein upregulation was linked to a subgroup of primary and
metastatic patients with more aggressive disease, and exhibited a
notable overlap of genes involved in mitotic regulation, these
results further support the hypothesis of TOP2A as a
biomarker for the early identification of patients with increased
metastatic potential that may benefit from adjuvant or neoadjuvant
targeted therapy approaches (26).
UBE2C serves as the key component in the ubiquitin
proteasome system by partnering with the anaphase-promoting complex
(APC/C). Upregulated UBE2C protein expression has been
reported in various types of human tumours (27). Mo et al (28) conducted an immunoassay to examine
209 breast cancer (BRCA) tissue samples and 53 normal tissue
samples, and found that UBE2C is highly expressed in BRCA.
Furthermore, the expression of UBE2C was positively
correlated with tumour size (24).
Wang et al (29) revealed
that the level of phosphorylated aurora kinase A (p-AURKA)
decreased markedly via the Wnt/β-catenin and PI3K/AKT signalling
pathways following the knockdown of UBE2C with a small
interfering RNA. The signalling pathway suppressed the occurrence
and development of gastric cancer, and their data suggested that
the activity of AURKA may be regulated by UBE2C via
modulation of the activity of APC/C (29), UBE2C may be a novel marker
in the diagnosis of gastric cancer (25); however, little is known about
UBE2C in seminoma. PTPRC is a member of the protein tyrosine
phosphatase (PTP) family. PTPs are signalling molecules that
regulate a variety of cellular processes, including cell growth,
differentiation, mitosis, and oncogenic transformation (30). Porcu et al (31) demonstrated that the downregulation
of CD45 (encoded by PTPRC) expression sensitizes T
cells to cytokine stimulation, as observed by increased JAK/STAT
signalling, whereas the overexpression of CD45 decreases
cytokine-induced signalling. In our study, PTPRC, which may
serve an important role in cytokine induction, was upregulated in
seminoma. The expression of MKI67 is strongly associated
with tumour cell proliferation and growth, and is widely employed
in routine pathological investigations as a proliferation marker
(32). It was revealed that the
expression of P53 and Wnt signalling correlated with that the
expression of MKI-67 in several types of cancer (33–36).
Downregulated MKI67 may be involved in cancer; in our study,
it was downregulated in seminoma. Five pivotal genes detected in
this study have been reported to be overexpressed in various human
cancers, are associated with their prognosis and are significantly
expressed in testicular seminoma tissues; but no significant
difference in prognosis was reported. The role of these genes in
TGCT is unclear; thus, further investigation is required.
Compared with normal testis samples, 11 DEMs were
acquired in GSE59520 in seminoma samples. In the present study,
hsa-miR-1182 was downregulated in seminoma samples. Zhou
et al (37) reported that
hsa-miR-1182 is dysregulated in bladder cancer tissues and
cell lines, in which functional assays were then performed, the
overexpression of miR-1182 significantly inhibits bladder cancer
cell proliferation, colony formation and invasion (37,38);
however, the effects of hsa-miR-1182 in seminoma are yet to
be determined. Hsa-miR-650 has been reported in many
cancers. For example, Zhou et al (39) observed that the expression of
miR-650 in tumour tissues had a positive association with OS.
Hsa-miR-650 inhibited cell growth and invasion in
vitro and in vivo (40,41).
Furthermore, miR-650 targeted AKT2 and suppressed the activation of
the AKT2/glycogen synthase kinase-3β/E-cadherin (39). Interestingly, Yang et al
(42) suggested that phosphatase
and tensin homolog/AKT signalling affects the expression of
TOP2A, reducing cell growth and inducing the apoptosis of
human breast cancer MCF-7 cells through ATP and caspase-3
signalling pathways (42). In our
study, TOP2A was identified as the target gene of
hsa-miR-650, and hsa-miR-650 was downregulated in
seminoma, while TOP2A was upregulated. Thus, we proposed
that in seminoma, the downregulation of hsa-miR-650 could
lead to the activation of the AKT pathway, upregulating
TOP2A to affect seminoma cell migration and proliferation.
At present, few studies have been conducted in the investigation of
hsa-miR-934. For hsa-miR-1204, Xu et al
(43) revealed that
hsa-miR-1204 expression was significantly correlated with
tumour size. The expression levels of hsa-miR-1204 and
glucose transporter-1 (GLUT-1) were significantly high in ovarian
cancer (OC) patients (43). The
expression levels of hsa-miR-1204 were positively correlated
with the expression levels of GLUT-1 in OC patients.
Hsa-miR-1204 overexpression significantly promoted GLUT-1
expression, glucose uptake and cell proliferation. In our study,
the target gene of hsa-miR-1204, CENPA, was revealed to
mostly function in kinetochores and regulate cell division. The
overexpression of CENPA is significantly related to colon
cancer and neoplastic germ cells (44,45).
Hsa-miR-1204 and its target gene CENPA were both
upregulated in seminoma; kinetochores are unique centromere
macromolecular protein structures that attach chromosomes to the
spindle for proper movement and segregation, during this process,
an increasing amount of ATP is required (46), and the overexpression of
hsa-miR-1204 can significantly promote GLUT-1 expression,
glucose uptake and ATP production (43). Thus, this pathway may affect the
expression of CENPA; however, the association between
CENPA and hsa-miR-1204 requires further
investigation. In addition, hsa-miR-661 has been reported in
non-small-cell lung cancer and OC (47,48).
Hoffman et al (49) found
that low miR-661 expression correlates with poor outcomes in BRCA
that typically express wild-type p53. In the present study, miR-661
was downregulated in seminoma. Of note, hsa-miR-1203 has
been reported to be dysregulated in prostate cancer, small cell
carcinoma of the oesophagus and oesophageal squamous cell carcinoma
(50–52). In addition, Prashad et al
(53) reported that miR-665
suppresses neuroblastoma tumorigenesis by inhibiting c-MYC and
suggested the potential of hsa-miR-665 as an
antineuroblastoma therapeutic factor. Dong et al (54) found that miR-665 was downregulated
in osteosarcoma tissues compared with nontumourous tissues, and the
expression of miR-665 was inversely associated with the expression
of Rab23 in the osteosarcoma tissues. These results suggest that
miR-665 could act as a tumour suppressor gene in the development of
osteosarcoma (54); it has also
been reported to be related to the Wnt/β-catenin signalling pathway
(55). In our study, miR-665 was
downregulated in seminoma, and the target gene MKI67 was also
associated with the Wnt/β-catenin signalling pathway (34). For hsa-miR-640, Li et
al (56) demonstrated that
miR-640 was downregulated in paclitaxel-resistant formalin-fixed
paraffin-embedded tumour samples; however, Zhou et al
(57) found that miR-640 is
related to the vascular endothelial growth factor receptor 2-mTOR
pathway. In the present study, hsa-miR-640 was downregulated
in seminoma, and its target gene, MKI67, was also associated with
mTOR (58).
In summary, TOP2A, MKI67, PTPRC and UBE2C were
revealed to be potentially associated with the PI3K/AKT and
Wnt/β-catenin signalling pathways, while hsa-miR-650 and
hsa-miR-665 were proposed to be linked the PI3K/AKT and
Wnt/β-catenin signalling pathways. Moreover, TOP2A and MKI67 were
strongly associated with the target genes of hsa-miR-650 and
hsa-miR-665, respectively.
At present, few bioinformatics analyses have been
conducted to investigate DEGs and DEMs in seminoma. Of the DEGs
screened in this study, only TOP2A and PTPRC have been reported in
seminoma (59,60), to the best of our knowledge.
Additionally, we screened the differential expression of genes from
array data and predicted target genes of DEMs which were proposed
to act together on the PI3K/AKT and Wnt/β-catenin signalling
pathways with a strong correlation (29,39).
The PI3K/AKT and Wnt/β-catenin signalling pathways participate in
the growth, invasion, and migration of cancer cells in a variety of
ways (61–63). TOP2A, MKI67, PTPRC and
UBE2C could be used as potential diagnostic biomarkers and
therapeutic molecular targets for seminoma. In addition, the main
treatment methods for seminoma in clinical practice are
chemotherapy and testicular resection, which undoubtedly cause
great physical burden to patients, particularly in individuals aged
15–24 years (7,64). Therefore, our findings of the
present study, including the genes identified, may serve as a basis
for exploring gene therapy for seminoma in the future.
In this study, 287 differential genes for testicular
seminoma were detected from the GEO database, and 5 pivotal genes
and 8 miRNAs were screened, all of which were significantly
differentially expressed in testicular cancer tissues. Among them,
we predicted the target genes of the miRNAs. TOP2A, MKI67, PTPRC
and UBE2C were determined to be associated with the PI3K/AKT and
Wnt/β-catenin signalling pathways, and hsa-miR-650 and
hsa-miR-665 were associated with the PI3K/AKT and
Wnt/β-catenin signalling pathways. Furthermore, TOP2A and MKI67
were strongly associated with the target genes of
hsa-miR-650 and hsa-miR-665, respectively. To the
best of our knowledge, hsa-miR-934 has not been
investigated. There may be certain associations that are yet to be
identified; the hub genes we reported could have notable impact on
cell proliferation and migration in testicular seminoma. In
addition, in patients with testicular seminoma, PTPRC
overexpression is an unfavourable prognostic factor, and further
studies are needed to verify our findings. The results of the
present study suggested that TOP2A, MKI67, CENPA, PTPRC, UBE2C,
hsa-miR-650, hsa-miR-665, hsa-miR-640, hsa-miR-1182,
hsa-miR-1203, hsa-miR-661 and hsa-miR-1204 may be
potential targets for seminoma therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasests generated and/or analyzed during the
current study are available from the Gene Expression Omnibus
repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15220,
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1818
and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59520.
Authors' contributions
KW and SZ made substantial contributions to the
conception of the present study. KW performed the primary
bioinformatics analysis and was a major contributor in writing the
manuscript; YC made substantial contributions to data analysis,
including the biological significance of hub genes and figure
editing. ZZ and MF were involved in the interpretation of the hub
genes data and edited the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pedersen MR, Rafaelsen SR, Møller H,
Vedsted P and Osther PJ: Testicular microlithiasis and testicular
cancer: Review of the literature. Int Urol Nephrol. 48:1079–1086.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith ZL, Werntz RP and Eggener SE:
Testicular cancer: Epidemiology, diagnosis, and management. Med
Clin North Am. 102:251–264. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ostrowski KA and Walsh TJ: Infertility
with testicular cancer. Urol Clin North Am. 42:409–420. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McNally RJ, Basta NO, Errington S, James
PW, Norman PD, Hale JP and Pearce MS: Socioeconomic patterning in
the incidence and survival of teenage and young adult men aged
between 15 and 24 years diagnosed with non-seminoma testicular
cancer in northern england. Urol Oncol. 33:506.e9–e14. 2015.
View Article : Google Scholar
|
|
5
|
Sharma P, Dhillon J and Sexton WJ:
Intratubular germ cell neoplasia of the testis, bilateral
testicular cancer, and aberrant histologies. Urol Clin North Am.
42:277–285. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baird DC, Meyers GJ and Hu JS: Testicular
cancer: Diagnosis and treatment. Am Fam Physician. 97:261–268.
2018.PubMed/NCBI
|
|
7
|
Kamel MH, Elfaramawi M, Jadhav S, Saafan
A, Raheem OA and Davis R: Insurance status and differences in
treatment and survival of testicular cancer patients. Urology.
87:140–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shen AH, Howell D, Edwards E, Warde P,
Matthew A and Jones JM: The experience of patients with early-stage
testicular cancer during the transition from active treatment to
follow-up surveillance. Urol Oncol. 34:168.e11–e20. 2016.
View Article : Google Scholar
|
|
9
|
Brand S, Williams H and Braybrooke J: How
has early testicular cancer affected your life? A study of sexual
function in men attending active surveillance for stage one
testicular cancer. Eur J Oncol Nurs. 19:278–281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
McBride D: Surveillance is as effective as
chemotherapy in stage 1 testicular cancer. ONS Connect.
29:102014.
|
|
11
|
Cheung HH, Lee TL, Davis AJ, Taft DH,
Rennert OM and Chan WY: Genome-wide DNA methylation profiling
reveals novel epigenetically regulated genes and non-coding RNAs in
human testicular cancer. Br J Cancer. 102:419–427. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Skotheim RI, Lind GE, Monni O, Nesland JM,
Abeler VM, Fosså SD, Duale N, Brunborg G, Kallioniemi O, Andrews PW
and Lothe RA: Differentiation of human embryonal carcinomas in
vitro and in vivo reveals expression profiles relevant to normal
development. Cancer Res. 65:5588–5598. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: CytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bender JL, Wiljer D, To MJ, Bedard PL,
Chung P, Jewett MA, Matthew A, Moore M, Warde P and Gospodarowicz
M: Testicular cancer survivors' supportive care needs and use of
online support: A cross-sectional survey. Support Care Cancer.
20:2737–2746. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Trabert B, Chen J, Devesa SS, Bray F and
McGlynn KA: International patterns and trends in testicular cancer
incidence, overall and by histologic subtype, 1973–2007. Andrology.
3:4–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lobo J, Costa AL, Vilela-Salgueiro B,
Rodrigues Â, Guimarães R, Cantante M, Lopes P, Antunes L, Jerónimo
C and Henrique R: Testicular germ cell tumors: Revisiting a series
in light of the new WHO classification and AJCC staging systems,
focusing on challenges for pathologists. Hum Pathol. 82:113–124.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Russell SS: Testicular cancer: Overview
and implications for health care providers. Urol Nurs. 34:172–176,
192. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Delgado JL, Hsieh CM, Chan NL and Hiasa H:
Topoisomerases as anticancer targets. Biochem J. 475:373–398. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Panvichian R, Tantiwetrueangdet A,
Angkathunyakul N and Leelaudomlipi S: TOP2A amplification and
overexpression in hepatocellular carcinoma tissues. Biomed Res Int.
2015:3816022015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Coleman LW, Perkins SL, Bronstein IB and
Holden JA: Expression of DNA toposiomerase I and DNA topoisomerase
II-alpha in testicular seminomas. Hum Pathol. 31:728–733. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sano K and Shuhin T: A study of
topoisomerase activity in human testicular cancers. Anticancer Res.
15:2117–2120. 1995.PubMed/NCBI
|
|
23
|
Meng H, Chen R, Li W and Xu L and Xu L:
Correlations of TOP2A gene aberrations and expression of
topoisomerase IIα protein and TOP2A mRNA expression in primary
breast cancer: A retrospective study of 86 cases using fluorescence
in situ hybridization and immunohistochemistry. Pathol Int.
62:391–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jensen JD, Knoop A, Ewertz M and Laenkholm
AV: ER, HER2, and TOP2A expression in primary tumor, synchronous
axillary nodes, and asynchronous metastases in breast cancer.
Breast Cancer Res Treat. 132:511–521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Konecny GE, Pauletti G, Untch M, Wang HJ,
Möbus V, Kuhn W, Thomssen C, Harbeck N, Wang L, Apple S, et al:
Association between HER2, TOP2A, and response to
anthracycline-based preoperative chemotherapy in high-risk primary
breast cancer. Breast Cancer Res Treat. 120:481–489. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Labbé DP, Sweeney CJ, Brown M, Galbo P,
Rosario S, Wadosky KM, Ku SY, Sjöström M, Alshalalfa M, Erho N, et
al: TOP2A and EZH2 provide early detection of an aggressive
prostate cancer subgroup. Clin Cancer Res. 23:7072–7083. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xie C, Powell C, Yao M, Wu J and Dong Q:
Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int
J Biochem Cell Biol. 47:113–117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mo CH, Gao L, Zhu XF, Wei KL, Zeng JJ,
Chen G and Feng ZB: The clinicopathological significance of UBE2C
in breast cancer: A study based on immunohistochemistry, microarray
and RNA-sequencing data. Cancer Cell Int. 17:832017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang R, Song Y, Liu X, Wang Q, Wang Y, Li
L, Kang C and Zhang Q: UBE2C induces EMT through Wnt/β-catenin and
PI3K/Akt signaling pathways by regulating phosphorylation levels of
Aurora-A. Int J Oncol. 50:1116–1126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vang T, Miletic AV, Arimura Y, Tautz L,
Rickert RC and Mustelin T: Protein tyrosine phosphatases in
autoimmunity. Annu Rev Immunol. 26:29–55. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Porcu M, Kleppe M, Gianfelici V, Geerdens
E, De Keersmaecker K, Tartaglia M, Foà R, Soulier J, Cauwelier B,
Uyttebroeck A, et al: Mutation of the receptor tyrosine phosphatase
PTPRC (CD45) in T-cell acute lymphoblastic leukemia. Blood.
119:4476–4479. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Menon SS, Guruvayoorappan C, Sakthivel KM
and Rasmi RR: Ki-67 protein as a tumour proliferation marker. Clin
Chim Acta. 491:39–45. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang CK, Yu TD, Han CY, Qin W, Liao XW, Yu
L, Liu XG, Zhu GZ, Su H, Lu SC, et al: Genome-wide association
study of MKI67 expression and its clinical implications in
HBV-related hepatocellular carcinoma in Southern China. Cell
Physiol Biochem. 42:1342–1357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bleckmann A, Conradi LC, Menck K, Schmick
NA, Schubert A, Rietkötter E, Arackal J, Middel P, Schambony A,
Liersch T, et al: β-catenin-independent WNT signaling and Ki67 in
contrast to the estrogen receptor status are prognostic and
associated with poor prognosis in breast cancer liver metastases.
Clin Exp Metastasis. 33:309–323. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li LT, Jiang G, Chen Q and Zheng JN: Ki67
is a promising molecular target in the diagnosis of cancer
(review). Mol Med Rep. 11:1566–1572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gallegos I, Valdevenito JP, Miranda R and
Fernandez C: Immunohistochemistry expression of P53, Ki67, CD30,
and CD117 and presence of clinical metastasis at diagnosis of
testicular seminoma. Appl Immunohistochem Mol Morphol. 19:147–152.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhou J, Dai W and Song J: miR-1182
inhibits growth and mediates the chemosensitivity of bladder cancer
by targeting hTERT. Biochem Biophys Res Commun. 470:445–452. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang T, Zhao D, Wang Q, Yu X, Cui Y, Guo
L and Lu SH: MicroRNA-1322 regulates ECRG2 allele specifically and
acts as a potential biomarker in patients with esophageal squamous
cell carcinoma. Mol Carcinog. 52:581–590. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou C, Cui F, Li J, Wang D, Wei Y, Wu Y,
Wang J, Zhu H and Wang S: MiR-650 represses high-risk
non-metastatic colorectal cancer progression via inhibition of
AKT2/GSK3β/E-cadherin pathway. Oncotarget. 8:49534–49547.
2017.PubMed/NCBI
|
|
40
|
Ningning S, Libo S, Chuanbin W, Haijiang S
and Qing Z: MiR-650 regulates the proliferation, migration and
invasion of human oral cancer by targeting growth factor
independent 1 (Gfi1). Biochimie. 156:69–78. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu X, Chen H, Zhang Q, Xu J, Shi Q and
Wang M: MiR-650 inhibits proliferation, migration and invasion of
rheumatoid arthritis synovial fibroblasts by targeting AKT2. Biomed
Pharmacother. 88:535–541. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang Z, Liu Y, Shi C, Zhang Y, Lv R, Zhang
R, Wang Q and Wang Y: Suppression of PTEN/AKT signaling decreases
the expression of TUBB3 and TOP2A with subsequent inhibition of
cell growth and induction of apoptosis in human breast cancer MCF-7
cells via ATP and caspase-3 signaling pathways. Oncol Rep.
37:1011–1019. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu J, Gu X, Yang X and Meng Y: MiR-1204
promotes ovarian squamous cell carcinoma growth by increasing
glucose uptake. Biosci Biotechnol Biochem. 1–6. 2018.(Epub ahead of
print).
|
|
44
|
Biermann K, Heukamp LC, Steger K, Zhou H,
Franke FE, Guetgemann I, Sonnack V, Brehm R, Berg J, Bastian PJ, et
al: Gene expression profiling identifies new biological markers of
neoplastic germ cells. Anticancer Res. 27:3091–3100.
2007.PubMed/NCBI
|
|
45
|
Liu R, Zhang W, Liu ZQ and Zhou HH:
Associating transcriptional modules with colon cancer survival
through weighted gene co-expression network analysis. Bmc Genomics.
18:3612017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Collins CM, Malacrida B, Burke C, Kiely PA
and Dunleavy EM: ATP synthase F1 subunits recruited to
centromeres by CENP-A are required for male meiosis. Nat Commun.
9:27022018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu F, Cai Y, Rong X, Chen J, Zheng D,
Chen L, Zhang J, Luo R, Zhao P and Ruan J: MiR-661 promotes tumor
invasion and metastasis by directly inhibiting RB1 in non small
cell lung cancer. Mol Cancer. 16:1222017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhu T, Yuan J, Wang Y, Gong C, Xie Y and
Li H: MiR-661 contributed to cell proliferation of human ovarian
cancer cells by repressing INPP5J expression. Biomed Pharmacother.
75:123–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hoffman Y, Bublik DR, Pilpel Y and Oren M:
miR-661 downregulates both Mdm2 and Mdm4 to activate p53. Cell
Death Differ. 21:302–309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Haldrup C, Kosaka N, Ochiya T, Borre M,
Høyer S, Orntoft TF and Sorensen KD: Profiling of circulating
microRNAs for prostate cancer biomarker discovery. Drug Deliv
Transl Res. 4:19–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Okumura T, Shimada Y, Omura T, Hirano K,
Nagata T and Tsukada K: MicroRNA profiles to predict postoperative
prognosis in patients with small cell carcinoma of the esophagus.
Anticancer Res. 35:719–727. 2015.PubMed/NCBI
|
|
52
|
Okumura T, Kojima H, Miwa T, Sekine S,
Hashimoto I, Hojo S, Nagata T and Shimada Y: The expression of
microRNA 574-3p as a predictor of postoperative outcome in patients
with esophageal squamous cell carcinoma. World J Surg Oncol.
14:2282016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Prashad N: miR-665 targets c-MYC and HDAC8
to inhibit murine neuroblastoma cell growth. Oncotarget.
9:33186–33201. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Dong C, Du Q, Wang Z, Wang Y, Wu S and
Wang A: MicroRNA-665 suppressed the invasion and metastasis of
osteosarcoma by directly inhibiting RAB23. Am J Transl Res.
8:4975–4981. 2016.PubMed/NCBI
|
|
55
|
Li K, Pan J, Wang J, Liu F and Wang L:
MiR-665 regulates VSMCs proliferation via targeting FGF9 and MEF2D
and modulating activities of Wnt/β-catenin signaling. Am J Transl
Res. 9:4402–4414. 2017.PubMed/NCBI
|
|
56
|
Li X, Lu Y, Chen Y, Lu W and Xie X:
MicroRNA profile of paclitaxel-resistant serous ovarian carcinoma
based on formalin-fixed paraffin-embedded samples. Bmc Cancer.
13:2162013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhou Y, Li XH, Zhang CC, Wang MJ, Xue WL,
Wu DD, Ma FF, Li WW, Tao BB and Zhu YC: Hydrogen sulfide promotes
angiogenesis by downregulating miR-640 via the VEGFR2/mTOR pathway.
Am J Physiol Cell Physiol. 310:C305–C317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yanai A, Inoue N, Yagi T, Nishimukai A,
Miyagawa Y, Murase K, Imamura M, Enomoto Y, Takatsuka Y, Watanabe
T, et al: Activation of mTOR/S6K But Not MAPK pathways might be
associated with high Ki-67, ER(+), and HER2(−) breast cancer. Clin
Breast Cancer. 15:197–203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dimov ND, Zynger DL, Luan C, Kozlowski JM
and Yang XJ: Topoisomerase II alpha expression in testicular germ
cell tumors. Urology. 69:955–961. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Bo H, Cao K, Tang R, Zhang H, Gong Z, Liu
Z, Liu J, Li J and Fan L: A network-based approach to identify DNA
methylation and its involved molecular pathways in testicular germ
cell tumors. J Cancer. 10:893–902. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yang Q, Jiang W and Hou P: Emerging role
of PI3K/AKT in tumor-related epigenetic regulation. Semin Cancer
Biol. Apr 2–2019.(Epub ahead of print). View Article : Google Scholar
|
|
62
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fruman DA and Rommel C: PI3K and cancer:
Lessons, challenges and opportunities. Nat Rev Drug Discov.
13:140–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Rajpert-De Meyts E, McGlynn KA, Okamoto K,
Jewett MA and Bokemeyer C: Testicular germ cell tumours. Lancet.
387:1762–1774. 2016. View Article : Google Scholar : PubMed/NCBI
|